Publications
Mondrian Abstraction and Language Model Embeddings for Differential Pathway Analysis
Fuad Al Abir & Jake Chen
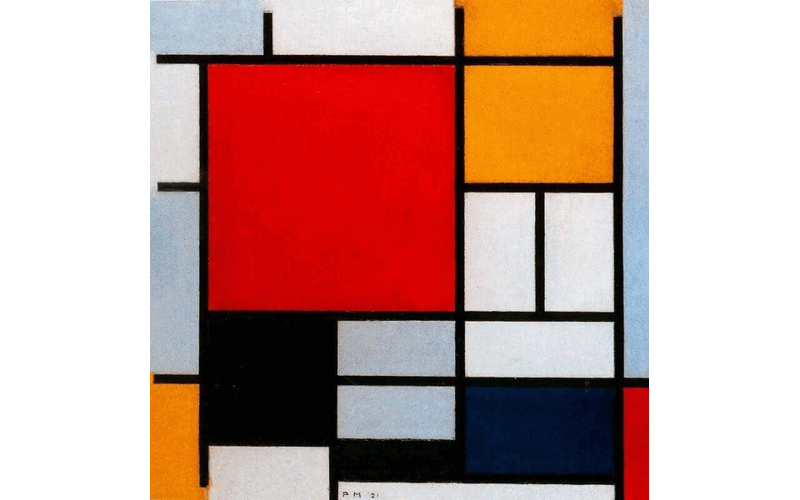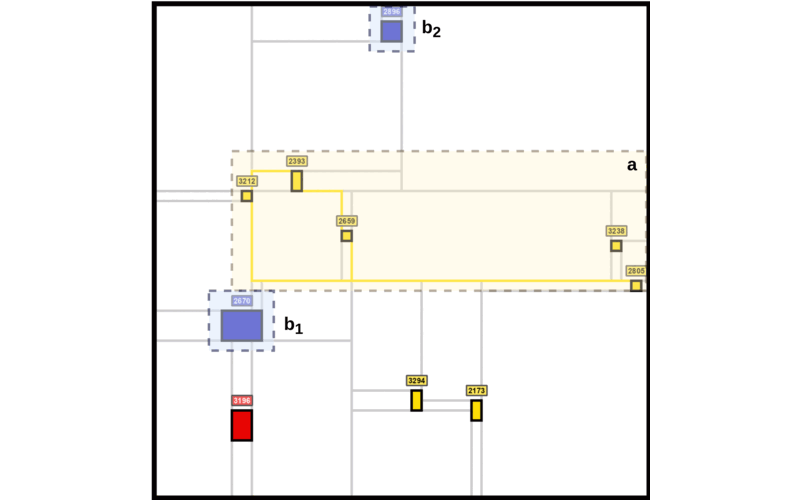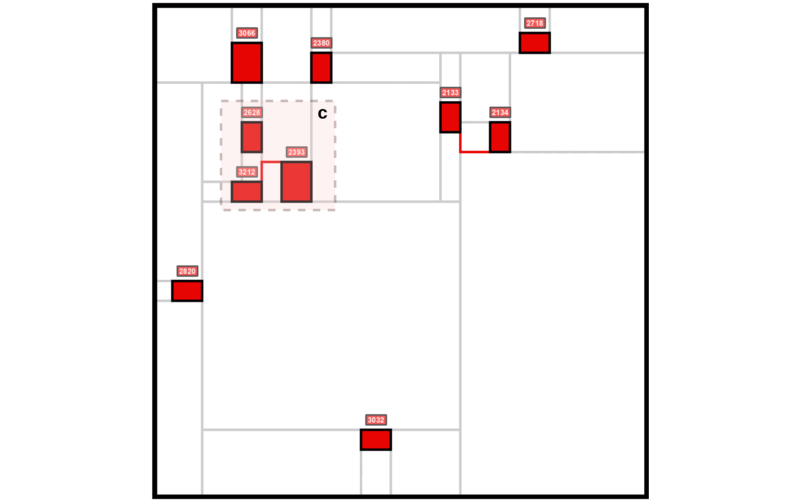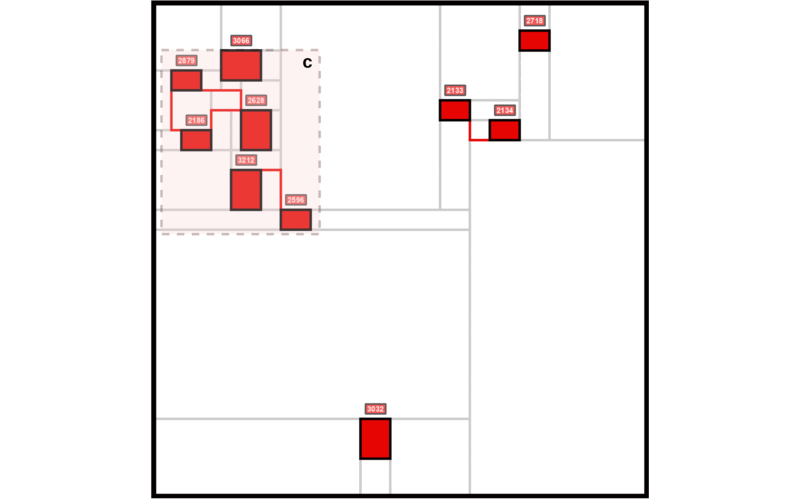
The Mondrian Map is a new visualization tool designed to simplify the complexity of biological networks by presenting pathways as structured, intuitive squares inspired by Mondrian’s abstract art. Our key contribution is an approach that retains critical relationships between pathways using language model embeddings while offering a visually accessible format, making it easier to interpret molecular dynamics and uncover insights in contexts like glioblastoma progression.
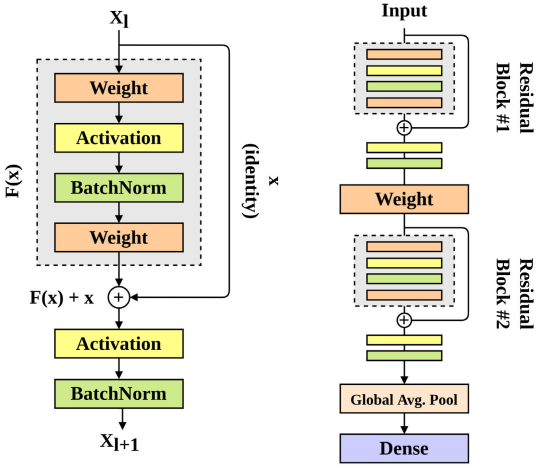
Gait Recognition with Wearable Sensors using Modified Residual Block-based Lightweight CNN
Fuad Al Abir*, Md Al Mehedi Hasan*, Md Al Siam, Jungpil Shin (* denotes equal contribution)
We tackle the challenge of gait recognition on low-power wearable devices by introducing an efficient deep learning model that maintains high prediction accuracy while minimizing model complexity. Our approach reduces model parameters and memory consumption by over 85% compared to existing methods, making it well-suited for wearable applications.

Deep Learning Based Air-Writing Recognition with the Choice of Proper Interpolation Technique
Fuad Al Abir, Md Al Siam, Abu Sayeed, Md Al Mehedi Hasan, Jungpil Shin
We address the challenge of recognizing air-writing gestures using smart-bands, focusing on minimizing signal duration variability without data loss. Our core contribution lies in employing effective interpolation techniques and developing a user-independent prediction model, making air-writing recognition more reliable and adaptable for real-world use, while outperforming existing methods across multiple datasets.
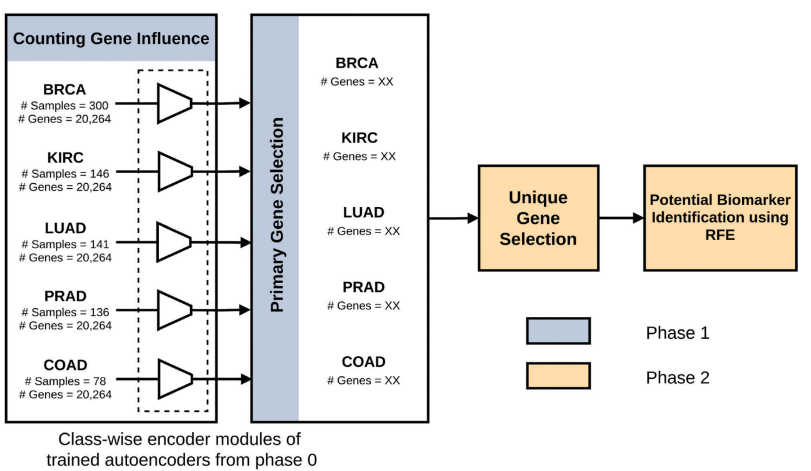
Biomarker Identification by Reversing the Learning Mechanism of Autoencoder and Recursive Feature Elimination
Fuad Al Abir, S M Shovan, Md Al Mehedi Hasan, Abu Sayeed, Jungpil Shin
We've developed a novel autoencoder-based method to enhance the identification of cancer biomarkers from RNA-Seq data, reversing traditional encoder learning mechanisms for more precise detection. Our methodology not only introduces a set of 17 highly potent biomarkers but also incorporates explainable AI techniques to clarify gene influences, significantly outperforming existing approaches in multiple diagnostic tasks.
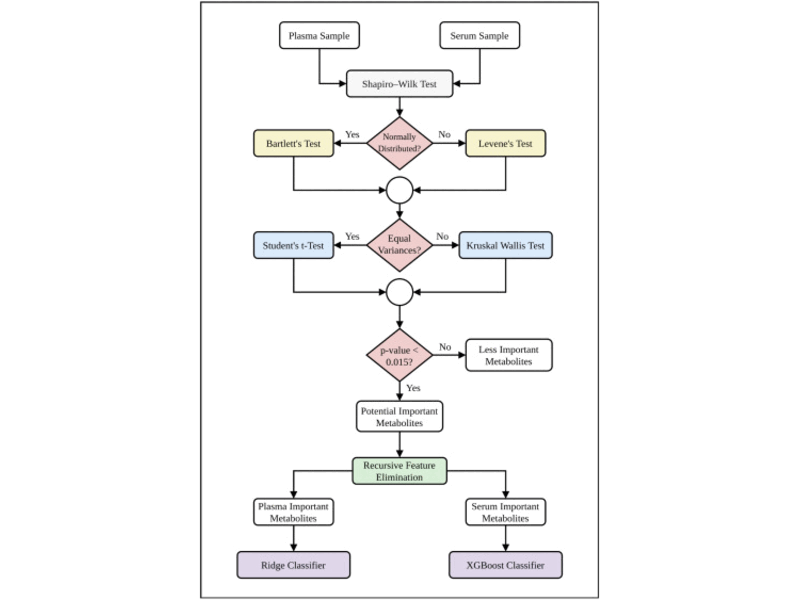
Most Dominant Metabolomic Biomarkers Identification for Lung Cancer
Utshab Kumar Ghosh, Fuad Al Abir, Nahian Rifaat, S M Shovan, Abu Sayeed, Md Al Mehedi Hasan
We address the challenge of identifying critical metabolomic biomarkers for early lung cancer prediction using blood samples. Our core contribution is a rigorous two-phase approach that integrates statistical tests and machine learning techniques, achieving exceptional prediction accuracy while highlighting dominant metabolites, offering valuable insights for advancing cancer diagnostics.
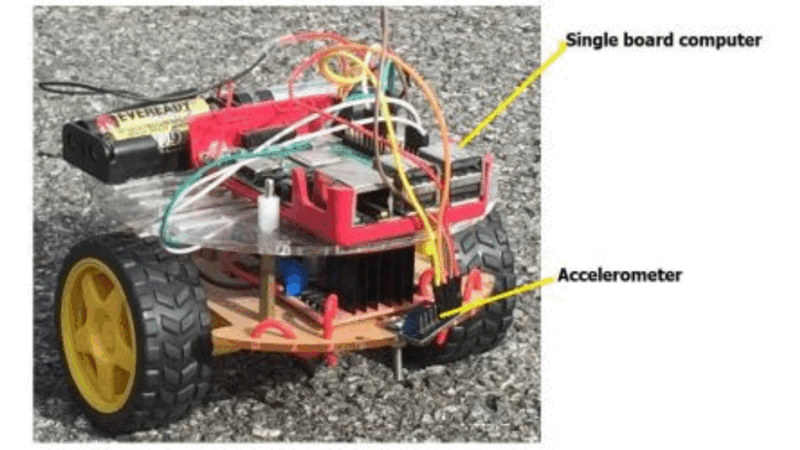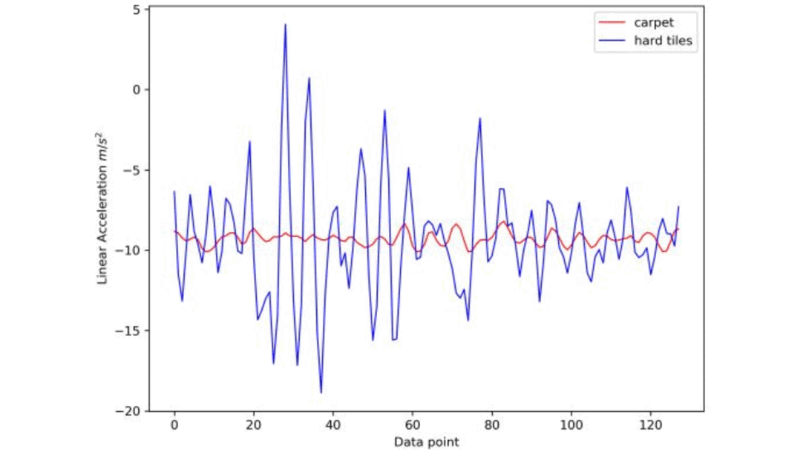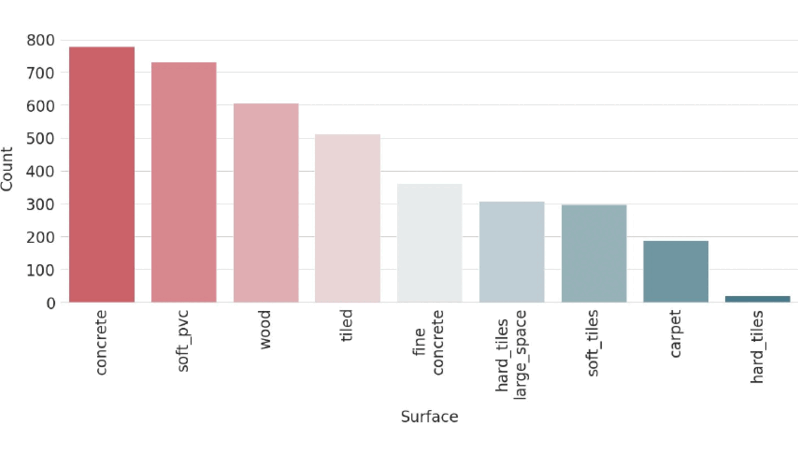
Surface Type Classification for Autonomous Robots Using Temporal, Statistical and Spectral Feature Extraction and Selection
Md Al Mehedi Hasan, Fuad Al Abir, Jungpil Shin
The challenge of real-time surface recognition for autonomous robots navigating complex indoor environments was addressed in this study. Our core contribution is an extensive feature extraction and classification method for time-series data that achieves state-of-the-art accuracy with minimal inference time, ensuring safe and efficient robot movement.
Current Projects
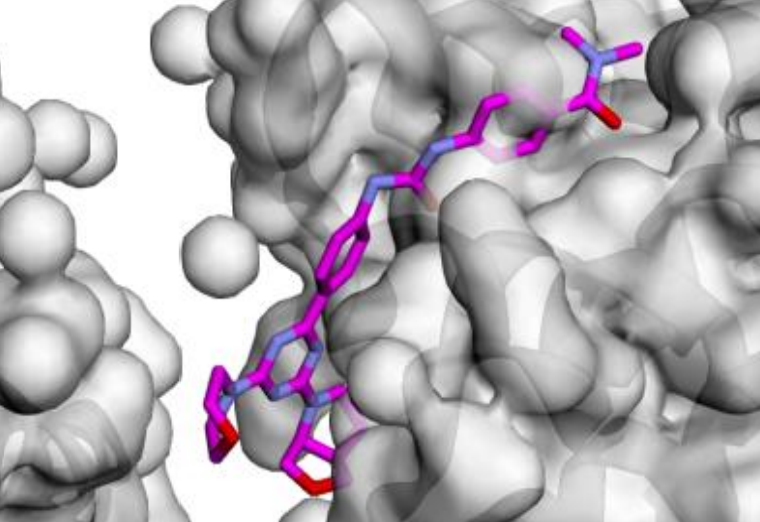
Guiding Chemical Language Models for Potent Breast Cancer Ligand Generation
Our research utilizes Chemical Language Models (CLMs) trained on SMILES for guided ligand generation for multiple targets, specifically focusing on breast cancer targets like mTOR and PI3K-α.
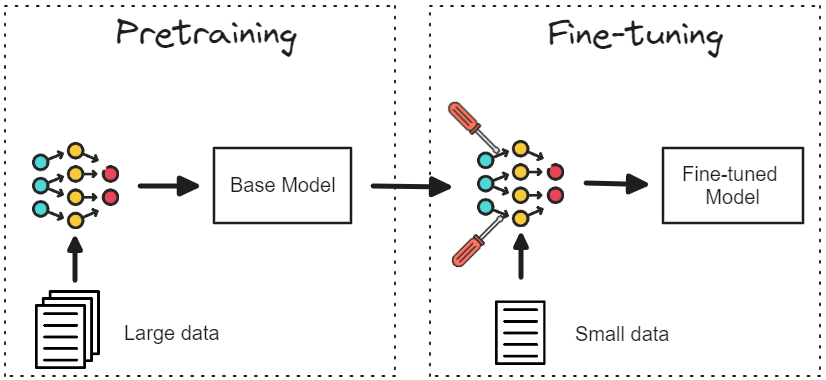
TherapMind: Progressive Curriculum Tuning of Large Language Models for Therapeutics
We introduce a curriculum-based learning framework designed to fine-tune Large Language Models (LLMs) for therapeutic applications, utilizing SMILES molecular representations and protein sequence data. This progressive task complexity enables LLMs to enhance their understanding of chemical and biological domains, overcoming limitations in existing LLM fine-tuning approaches.
Past Projects
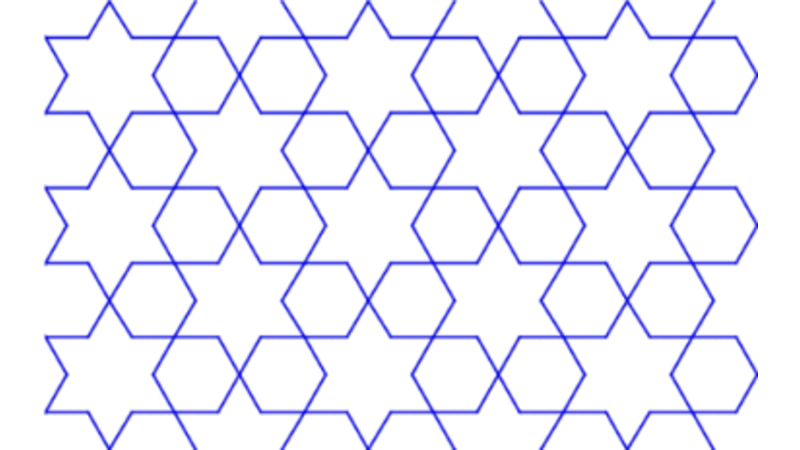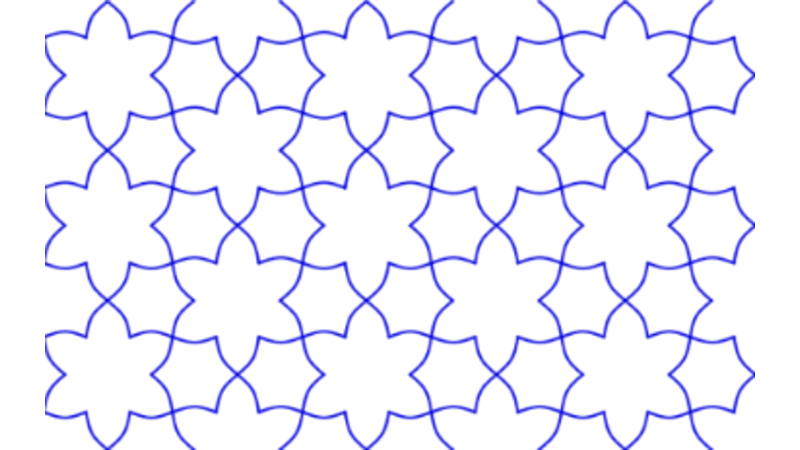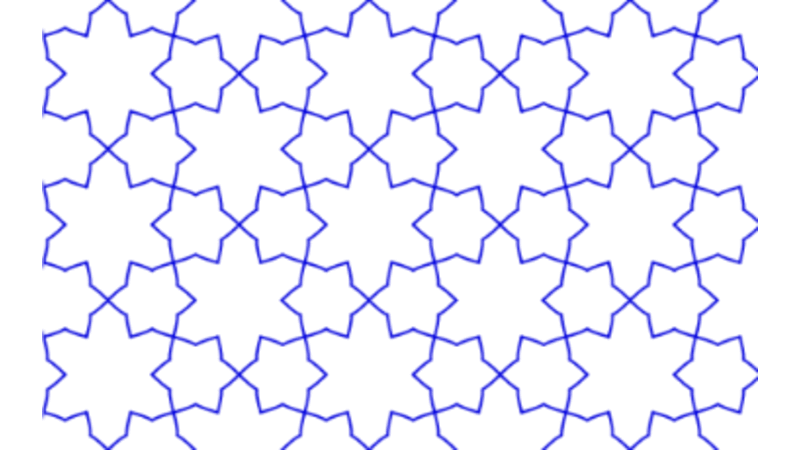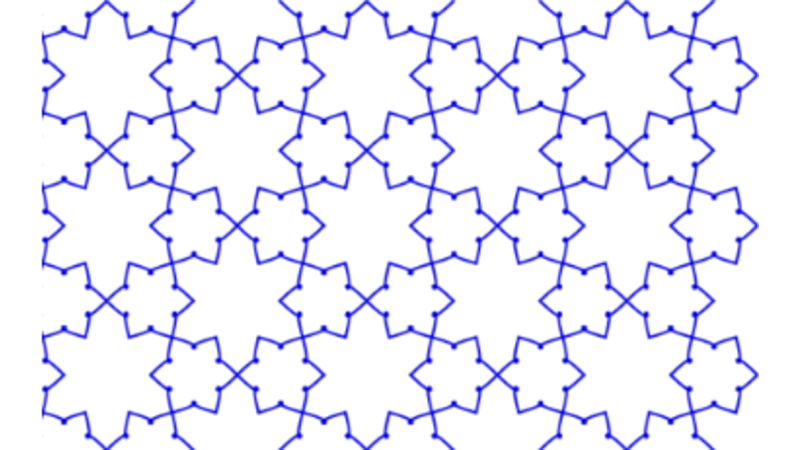
Islamic Geometric Patterns (IGPs) with Catmull-Rom Splines
This project focuses on the development of interactive software designed to generate Islamic Geometric Patterns (IGPs) using tesselation techniques. Leveraging the p5.js (processing) framework, the software facilitates user engagement with generative art, allowing for dynamic pattern creation. Additionally, a comprehensive literature review was conducted to explore existing methodologies in interactive computing and generative art, grounding the software's development in current academic and practical contexts.
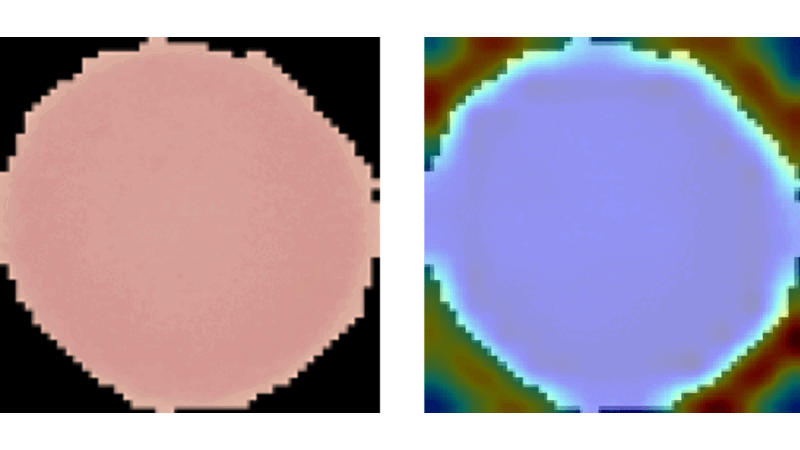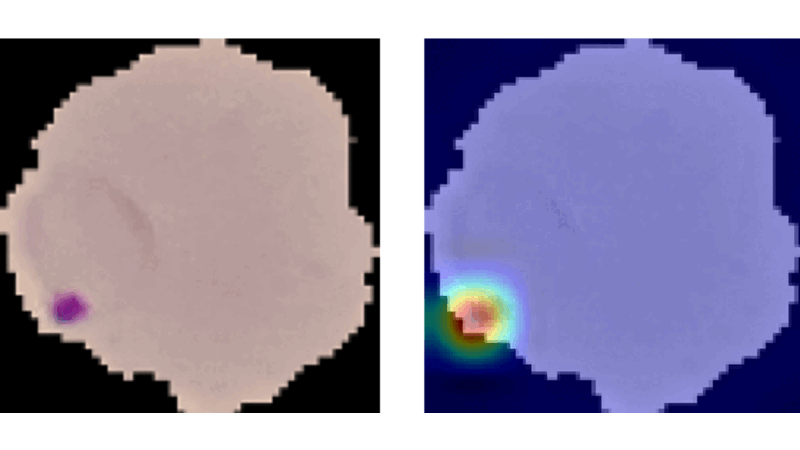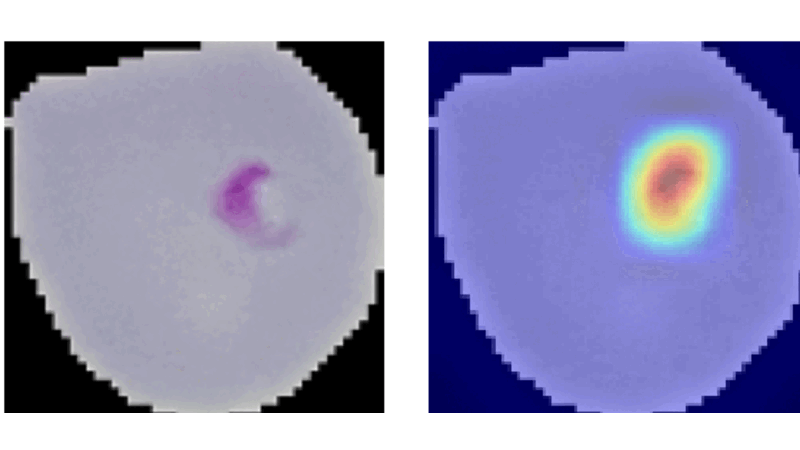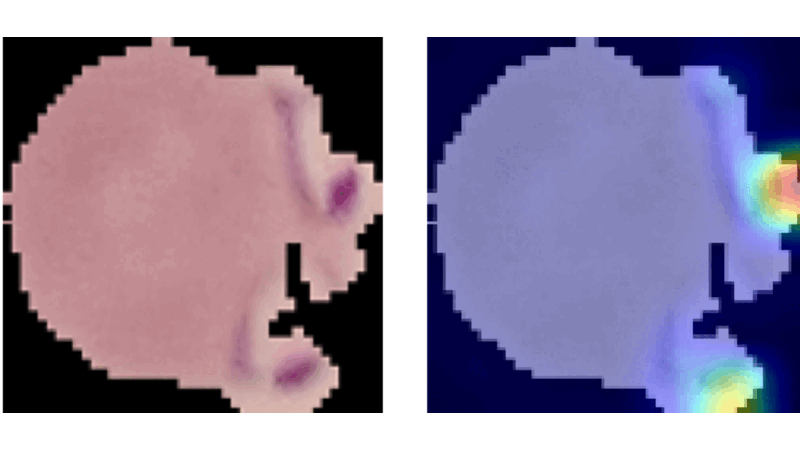
Explainable CNNs in Medical Imaging
This project aimed to enhance trust in AI applications within medicine by developing and evaluating advanced vision models on the NIH Malaria dataset. It involved creating a method similar to Grad-CAM from scratch, which aggregates propagation heatmaps to provide visual explanations of model decisions, thereby making AI decisions more transparent and understandable in medical contexts.